Section: New Results
Software Engineering
Participants : Jocelyn Gate, Stephane Redon.
We have continued the development of SAMSON, our open-architecture platform for modeling and simulation of nanosystems (SAMSON: Software for Adaptive Modeling and Simulation Of Nanosystems). The interface has been improved:
The visualization of the data graph has been improved. Users may now drag and drop models and parts between layers, as well as directly drag and drop files into SAMSON.
The software engineering process has been improved as well, in particular to help base and modules developers:
We have reorganized the file hierarchy so that modules can have associated data.
We have developed a system to build SAMSON automatically on virtual machines (e.g., ubuntu 12.04 32bit, ubuntu 12.04 64 bit, fedora 17 32 bit, etc.).
Tools have been created to let modules developers easily write new modules.
We have begun to develop a mechanism to make it easy to install and update SAMSON automatically.
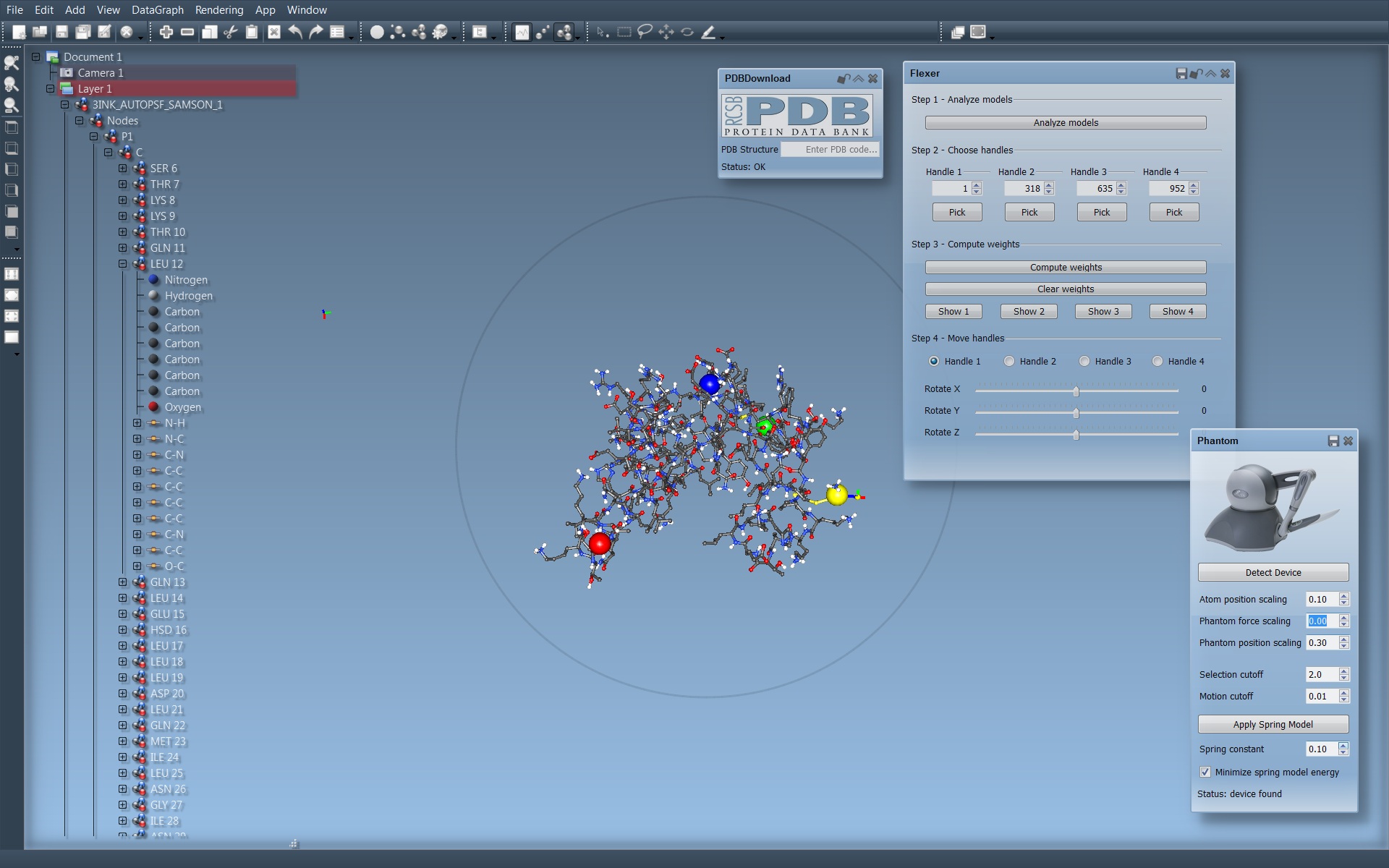
We have also developed several SAMSON apps to test various concepts, including scripting, manipulating molecules with haptic feedback, etc. Figure 12 shows the current user interface of SAMSON.
We have deposited the first version of SAMSON's code base at the APP ("Agence de Protection des Programmes").